

Knepper Laboratory
CPhos
(a program to identify evolutionarily conserved phosphorylation sites)
Description
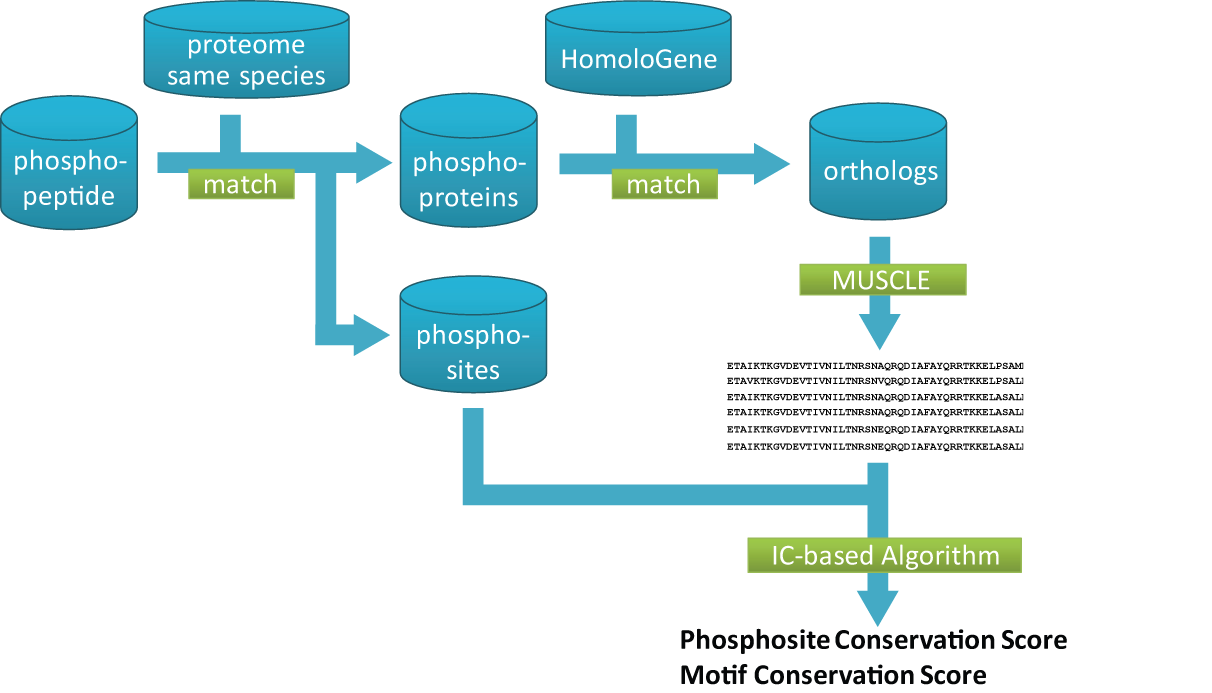
CPhos utilizes an information theory-based algorithm to assess the conservation of phosphorylation sites among species.
A conservation established from this approach can be used to potentially assess the functional significance of a particular phosphorylation site.
Download standalone versions of CPhos:
CPhos algorithm was developed by Boyang Zhao, Trairak Pisitkun, Jason D. Hoffert, Mark A. Knepper, and Fahad Saeed.
If you use this software please cite:
Zhao, B., Pisitkun, T., Hoffert, J. D., Knepper, M. A. and Saeed, F. (2012), CPhos: A program to calculate and visualize evolutionarily conserved functional phosphorylation sites. Proteomics, 12: 3299-3303. Abstract and link to full text
Please read the Frequently Asked Questions(FAQ) to solve common problems. Contact us if you have questions not answerable by the FAQ page or want to report bugs.
Instructions
- Select the species for phosphopeptides to be analyzed.
- Prepare an input text file containing a list of phosphopeptides with an asterisk symbol (*) indicated on the right-hand side of each phosphorylation site.
The maximum limit of input is 300 phosphopeptides. For a larger number of input phosphopeptides, please use standalone versions of CPhos (see download links below).
- Choose the prepared Enter input text file.
- Press "Submit" to run program.
- Upon completion, results will be displayed in the same window. Additional result data and raw data files can be downloaded via the link provided in the results page.
- To download results, click on "Download data" in the results page. The phosphopeptides and corresponding phosphoprotein, phosphosites, and conservation scores are summarized in the "phosphopeptide.txt", "phosphoproptein.txt", "phosphosite.txt", and "cscores.txt" files.
- To retrieve the raw outputs (i.e. FASTA file (.fasta), sequence alignments (.aln), and phylogenetic tree (.ph)), click on "Download data" and then "rawoutputs". The files are named as the protein's HomoloGene ID (HID), which can be found in the results in step 6.
Example test input (containing real and fake phosphopeptides)
SPQS*LPRGS*K
ERRGDNLGS*P
LPPLVIPZS*E
SDF
LPPLVIPPS*E
Algorithm and Program Work flow
The general work flow of CPhos is illustrated below.
Go to ESBL Main Page




